Rhodium »
PDB 165d-8oqd »
4kii »
Rhodium in PDB 4kii: Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide
Protein crystallography data
The structure of Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide, PDB code: 4kii
was solved by
M.V.Cherrier,
P.Amara,
S.Engilberge,
J.C.Fontecilla-Camps,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.86 / 1.85 |
| Space group | P 32 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.950, 52.950, 110.680, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.7 / 22 |
Rhodium Binding Sites:
The binding sites of Rhodium atom in the Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide
(pdb code 4kii). This binding sites where shown within
5.0 Angstroms radius around Rhodium atom.
In total only one binding site of Rhodium was determined in the Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide, PDB code: 4kii:
In total only one binding site of Rhodium was determined in the Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide, PDB code: 4kii:
Rhodium binding site 1 out of 1 in 4kii
Go back to
Rhodium binding site 1 out
of 1 in the Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide
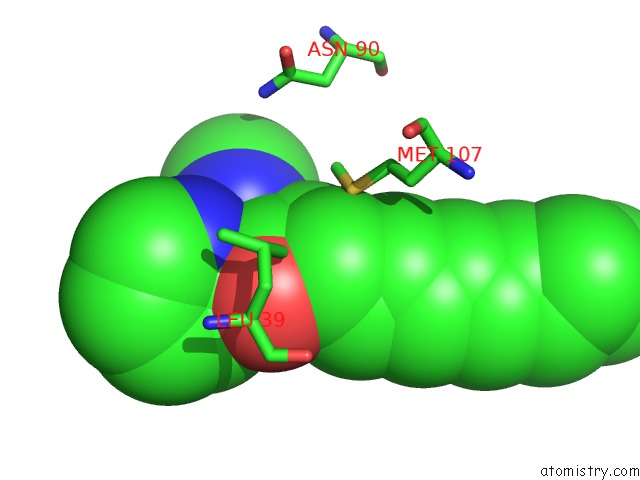
Mono view
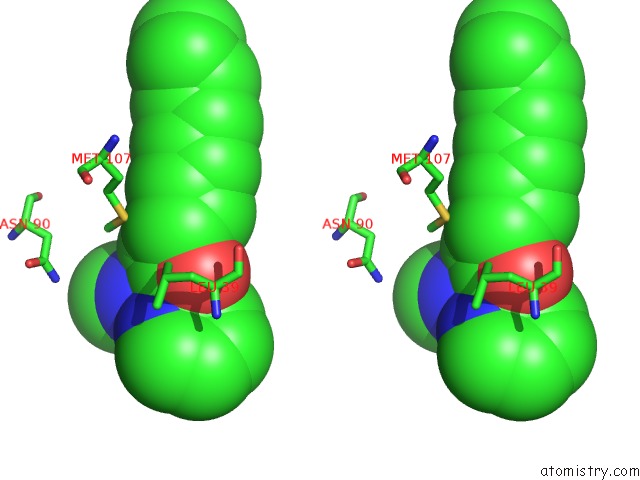
Stereo pair view

Mono view

Stereo pair view
A full contact list of Rhodium with other atoms in the Rh binding
site number 1 of Beta-Lactoglobulin in Complex with Cp*Rh(III)Cl N,N-Di(Pyridin-2-Yl) Dodecanamide within 5.0Å range:
|
Reference:
M.V.Cherrier,
P.Amara,
S.Engilberge,
A.Chevalley,
M.Salmain,
J.C.Fontecilla-Camps.
Structural Basis For Enantioselectivity in the Transfer Hydrogenation of A Ketone Catalyzed By An Artificial Metalloenzyme Eur.J.Inorg.Chem. V. 21 3596 2013.
ISSN: ISSN 1434-1948
DOI: 10.1002/EJIC.201300592
Page generated: Tue Aug 19 00:41:09 2025
ISSN: ISSN 1434-1948
DOI: 10.1002/EJIC.201300592
Last articles
Sr in 3BNQSr in 2QJY
Sr in 2X53
Sr in 2SPT
Sr in 2XRM
Sr in 2WOH
Sr in 2RIO
Sr in 2PN4
Sr in 2QJK
Sr in 2QJP